
Laboratory of Choice
Children are not things to be molded but are people to be unfolded.
Science-first leaders whose compassion for healthcare providers and patients is incomparable.
Curabitur arcu erat, accumsan id imperdiet et, porttitor at sem. Nulla porttitor accumsan tincidunt.
Comprehensive Gastro-Intestinal Solutions
Comprehensive Gastro-Intestinal Solutions
Comprehensive Gastro-Intestinal Solutions
Barrett’s Esophagus FISH
Barrett’s Esophagus (BE) Facts
BE is estimated to affect 10% to 12% of patients with chronic gastroesophageal reflux disease1. Patients with Barrett’s Esophagus (BE) are approximately 125 times more likely to develop Esophageal Adenocarcinoma (EA) than the general population2. The evolution to cancer from BE is generally accepted to be a multistep progression as follows:
- Intestinal Metaplasia (IM) without dysplasia
- Low-grade dysplasia (LGD)
- High-grade dysplasia (HGD) and Esophageal Adenocarcinoma (EA)
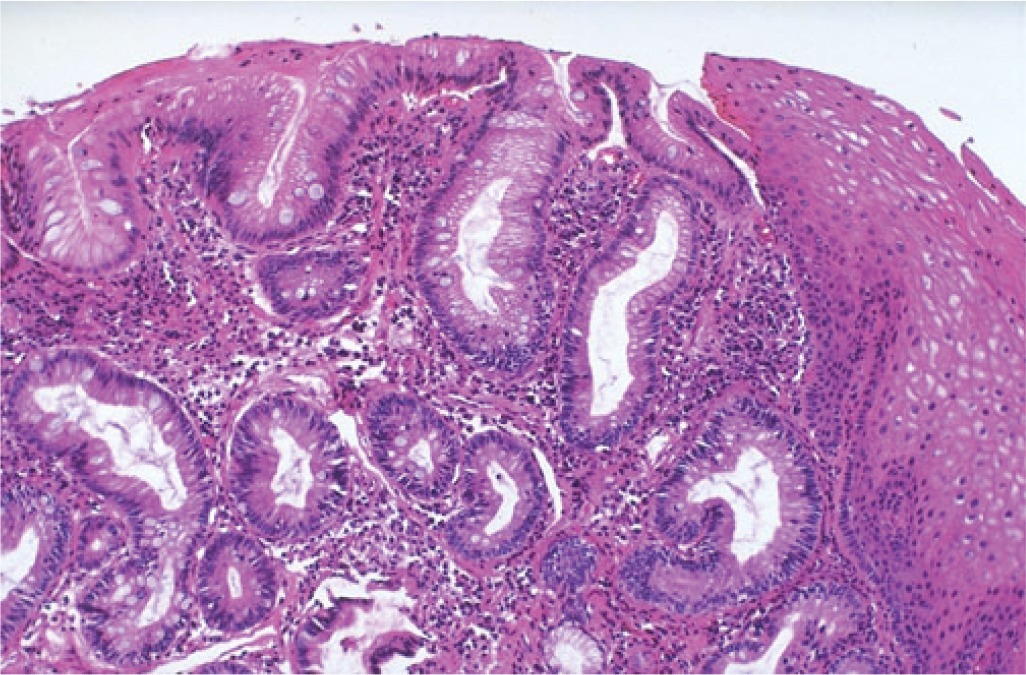
Challenges with BE Testing
The recommended surveillance method of 4 quadrant biopsies every 1-2 cm of the affected mucosa has several shortcomings which include: inadequate sampling of the affected tissue and the inability of the traditional H&E stains to show a number of genetic alterations reported in BE as IM progresses to EA.
Numerous studies have shown an increase in a number of genetic alterations as IM progresses to EA(3-7).
Premalignant lesions such as LGD have been shown to be diploid or near diploid with frequent alterations in P16 & P53(6-10).
Genesis Barrett’s Esophagus FISH Test
Genesis Lab’s Barrett’s FISH test deploys a FISH probe set that was developed specifically for detection of genetic abnormalities found in BE patients who are more likely to progress to HGD & EA even if no abnormalities are seen on traditional biopsies.
The probe set consists of directly labeled DNA probes to 8Q24 (C-MYC), 9p21 (p16), 17q12 (HER2) and 20q13.
Cytology specimens collected via endoscopic brushing result in more comprehensive and wide area sampling than biopsies alone. FISH testing is performed on the cytology sample as an adjunct to the four quadrant biopsies.
Studies have shown progression to HGD & EA in spite of no dysplasia being seen on traditional biopsies within 33 months of significant genetic abnormalities e.g. if polysomy are seen on the FISH testing.
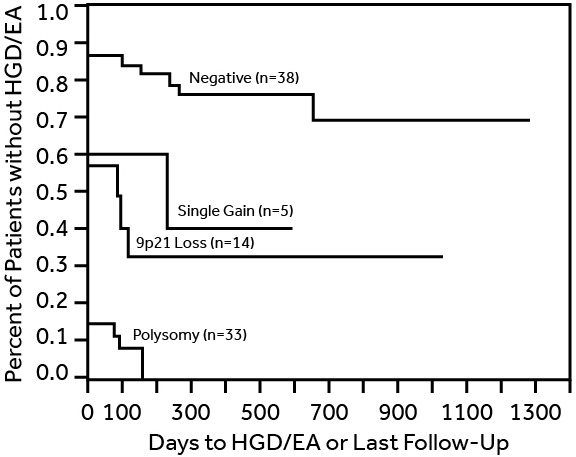
Genesis HGB/EA Chart
Fig. 1 Kaplan-Meier analysis demonstrates differences in time to HGD and or EA for patients based upon the FISH abnormalities found in initial brushing specimen11.
Addition of Genesis BE FISH testing would lead to identification of more high risk cases who are likely to progress to HGD/EA prior to their next recommended surveillance interval of 36 months . The surveillance interval should be shortened for patients noted to have significant genetic abnormalities identified on BE FISH testing.
Uses and Benefits
- Genesis Lab BE FISH test serves as an ancillary aid that adds the discriminatory power of genetic data to identify high risk patients more likely to develop HGD & EA.
- Helps in determining the prognosis of BE.
- Because Genesis Lab BE is performed on cytology brushings rather than biopsies, collection is rapid and a wider area of B.E. is sampled.
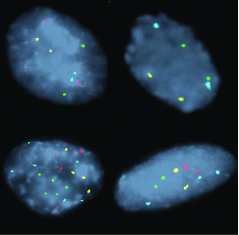
Fig2. Examples of FISH Patterns: (A) normal (2 signals of each of the 4 probes, (B) homozygous 9p21 loss (no red signals), © polysomy (2or more signals of 2 or more probes), (D) gain of 8q24 (greater than 2 aqua signals). 9p21 (red), 17q12 (green), 8q24(aqua), and 20q(gold)11.
Keep up-to-date with Genesis Labs.
46th Annual New York Course Conference
sean@rizco.com2022-12-02T15:58:47+00:00December 2, 2022|
Florida Digestive Diseases Update 2022
sean@rizco.com2022-12-02T16:49:20+00:00December 2, 2022|
Post ACG Conference
genesislabs222022-11-18T14:40:41+00:00November 17, 2022|
References
- Bonino JA, Sharma P. (2005). Barrett’s esophagus. Current Opinion Gastroenterology, (21),461-465.
- Cameron AJ, Ott BJ, Payne WS. (1985) The incidence of adenocarcinoma in columnar-lined (Barrett’s) esophagus. New England Journal of Medicine,(313),857-859.
- Riegman PH, Vissers KJ, Alers JC, et al.,(2001) Genomic alterations in malignant transformation of Barrett’s esophagus. Cancer Research, (61),3164-3170.
- Walch, A. K., Zitzelberger, H. F., Bruch, J., Keller, G., Angermeier, D., Aubele, M. M., . . . Werner, M. (2000). Chromosomal Imbalances in Barrett’s Adenocarcinoma and the Metaplasia-Dysplasia-Carcinoma Sequence. The American Journal of Pathology,156(2), 555-556. Retrieved from https://www.sciencedirect.com/science/article/pii/S0002944010647608
- Falk, G. W., Skacel, M., Gramlich, T. L., Casey, G., Goldblum, J. R., & Tubbs, R. R. (2004). Fluorescence in situ hybridization of cytologic specimens from Barrett’s esophagus: A pilot feasibility study. Gastrointestinal Endoscopy,60(2), 280-284. doi:10.1016/s0016-5107(04)01687-6
- Galipeau PC, Prevo LJ, Sanchez CA, Longton GM, Reid BJ.(1999). Clonal expansion and loss of heterozygosity at chromosomes 9p and 17p in premalignant esophageal (Barrett’s) tissue. JNCI: Journal of the National Cancer Institute, (91), 2087-2095.
- Brankley, S. M., Wang, K. K., Harwood, A. R., Miller, D. V., Legator, M. S., Lutzke, L. S., . . . Halling, K. C. (2006). The Development of a Fluorescence in Situ Hybridization Assay for the Detection of Dysplasia and Adenocarcinoma in Barrett’s Esophagus. The Journal of Molecular Diagnostics,8(2), 260-267. doi:10.2353/jmoldx.2006.050118
- Riegman, P. H., Burgart, L. J., Wang, K. K., Wink-Godschalk, J. C., Dinjens, W. N., Siersema, P. D., . . . Van Dekken, H. (2002). Allelic Imbalance of 7q32.3-q36.1 during Tumorigenesis in Barrett’s Esophagus. Cancer Research,62, 1531-1533. Retrieved from http://cancerres.aacrjournals.org/content/62/5/1531.full-text.pdf
- Barrett, M. T., Sanchez, C. A., Prevo, L. J., Wong, D. J., Galipeau, P. C., Paulson, T. G., . . . Reid, B. J. (1999). Evolution of neoplastic cell lineages in Barrett’s esophagus. Nature Genetics,22(1), 106-109. doi:10.1038/8816
- Wong, D., Prevo, L., Galipeau, P., Paulson, T., Blount, P., & Reid, B. (2001). Clonal cell populations with p16 lesions arise early and expand within Barrett’s metaplastic epithelium. Gastroenterology,120(5), 8284-82849. doi:10.1016/s0016-5085(01)80390-5
- Fritcher, E. G., Brankley, S. M., Kipp, B. R., Voss, J. S., Campion, M. B., Morrison, L. E., . . . Halling, K. C. (2008). A comparison of conventional cytology, DNA ploidy analysis, and fluorescence in situ hybridization for the detection of dysplasia and adenocarcinoma in patients with Barrett’s esophagus. Human Pathology,39(8), 1128-1135. doi:10.1016/j.humpath.2008.02.003


